Introducing EMMI: Where Experimentation Meets Machine Intelligence
Small molecule drug discovery is simultaneously an amazing human endeavor and a business ripe for disruption, as it transitions from a human intuition-driven journey to an AI-guided, reproducible process. Small molecules have transformed human health and make up 90% of medicines used globally, owing to their efficacy and ease of administration as pills. Despite this global impact, discovering new small molecule drugs is incredibly hard, with only a fraction of initial programs progressing to clinical testing (and most clinical testing ending in failure).
Successful discovery is a complex, multi-parameter optimization problem wherein on-target efficacy, off-target toxicity, distribution through the body, metabolism, excretion, and more must be balanced. Two key decisions in this process are which molecule(s) should you start from (hit finding), and which molecules should you synthesize and test in each iterative cycle to optimize the starting points (hit-to-lead and lead optimization) to reach a Development Candidate ready for clinical testing.
Every step requires synthesizing large amounts of data and making probabilistic choices as to what to do next experimentally, making it a natural fit for AI which can vastly outperform humans at these types of tasks. It has thus become common to suggest that training a general model on publicly available data or static datasets will solve drug discovery, but this overlooks the key limitation – by definition the hardest problems in human health have solutions that lie well outside of molecules that have already been tested, so models armed with only today’s commodity tools and datasets will struggle to address them. Indeed, many current “AI drugs” are strikingly similar to previous patents as the models perform well when tasked with finding a better solution to a previously addressed challenge.
Setting out in a different direction, Terray has built a chemistry-first, AI-native integrated platform for de novo solutions to unsolved challenges in medicine. Today, we are introducing that platform, EMMI, where Experimentation Meets Machine Intelligence.
EMMI Overview
EMMI integrates high-throughput experimental capabilities that rapidly produce precise, proprietary data at scale in data-poor areas of chemical space with a full-stack AI platform. Terray’s scientists interact with EMMI every single day to generate, predict, and select the right set of molecules for testing in every step of the drug discovery process. EMMI optimizes human-in-the-loop decision-making to drive each program at Terray and has delivered across a diverse portfolio of challenging drug discovery programs – both for Terray’s internal pipeline focused on immunology (which includes a co-development program with Odyssey Therapeutics), and in discovery partnerships with Calico, Bristol Myers Squibb, and Gilead.

The EMMI Platform
With the foundational platform in place, we’re incorporating deeper AI-reasoning and continually tightening the integration of AI-driven design and experimentation to move steadily towards a truly closed-loop process.
EMMI’s Experimentation Edge
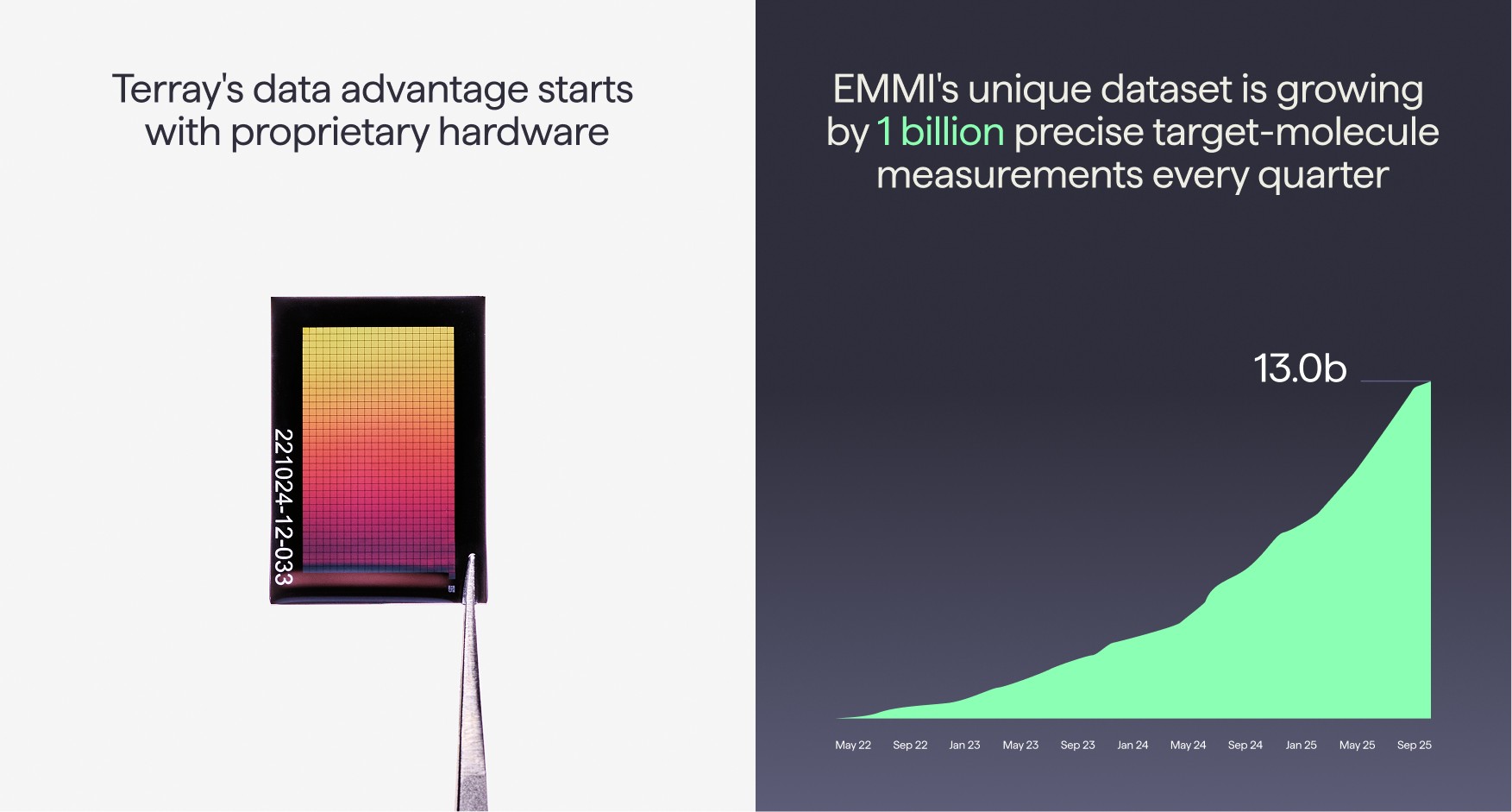
When we founded Terray, we knew that creating the right data that iterates at scale with precision would be the key to powering AI that could solve the previously unsolvable challenges in drug discovery. Existing high-throughput methods were either underpowered or had poor signal quality or lacked the ability to iterate, so we invented a novel ultra-dense microarray for measuring the interactions between millions of small molecules and targets of interest. This technology is used both to find novel hits with a growing collection of diversity libraries containing hundreds of millions of molecules and to rapidly explore promising areas of chemical space during hit-to-lead and lead optimization with custom-built focus libraries of millions of molecules. This proprietary core hardware has allowed us to explore the dark areas of chemical space with over 13 billion unique, precise measurements of target-molecule interactions. This is the largest global database of binding data for small molecules and grows at 1 billion measurements per quarter. Terray mines this database to follow up on the most promising molecules in Terray’s highly-automated experimental workflows, where a suite of purpose-built modular systems seamlessly operate in concert to synthesize and test molecules at scale.
EMMI’s Machine Intelligence Edge
The Machine Intelligence side of EMMI guides optimal decisions at each Design-Make-Test-Analyze (DMTA) cycle with a human-in-the-loop framework, as we work towards a full closed-loop process. The models we have developed are purpose-built to achieve this by addressing discovery needs arising in challenging small molecule drug discovery programs. These models fall into four distinct categories:
- Reason: Foundation and reasoning models that understand molecules and chemistry and guide scientists throughout the Generate -> Predict -> Select design workflow
- Generate: Propose new virtual molecules for further evaluation, typically inspired by an existing molecule
- Predict: Profile the properties of the generated molecules, such as potency and ADME
- Select: Identify the optimal number of molecules and which molecules to synthesize and test to achieve a multi-parameter objective
Reason: EMMI’s multi-modal foundation model of chemistry, COATI
To efficiently design molecules possessing a specified set of properties (also known as inverse design), AI tools must be able to speak the language of molecules. Traditional molecular representations like descriptors and fingerprints that have been used to encode structures as vectors are unsuitable for inverse design as they do not decode effectively once manipulated. In contrast, our chemical foundation model, COATI, encodes molecules in an invertible mathematical latent space. This transformation makes molecular structures accessible to AI models to manipulate and, most importantly, provides the language to translate the mathematical representations back into molecules. Conceptually, this is akin to a GPT for molecules. COATI is a multi-modal foundation model trained using contrastive learning, very similar to how OpenAI’s DALL-E is trained. Instead of contrasting images and text, we contrast various molecular representations like SMILES (molecules represented as strings), graphs, and conformers. And just like foundation models in other domains, we are continuously improving this model, moving from COATI1 to COATI3 in the last 2 years. COATI3 is trained on over one billion diverse molecules, is a 768-dimensional representation, and has a SMILES, 2D-graph, and 3D encoder with a SMILES decoder – allowing Terray to explore new areas of chemical space with precision. Developing this model is a complex process, and we have worked with NVIDIA to efficiently leverage their GPU compute infrastructure through DGX Cloud to train the family of COATI models - see our case study here: https://www.nvidia.com/en-us/customer-stories/generative-ai-for-small-molecule-drug-discovery Importantly, COATI is the underpinning of our Generate, Predict, and Select models, and the constant use and feedback further helps us identify ways to improve it.
For interested readers:
- See our COATI GitHub repository here: https://github.com/terraytherapeutics/COATI
- See our JCIM paper here: https://pubs.acs.org/doi/10.1021/acs.jcim.3c01753
Generate: EMMI’s generative models design property-optimized molecules with latent diffusion and reinforcement learning
The first step in molecular design is to propose or generate new virtual molecules for further evaluation, typically from an existing starting point. Depending on what is most efficient for the stage of program, this can be done with simple rule-based cheminformatics approaches or ML models that produce molecules with specified properties. These ML models leverage Terray’s proprietary datasets to learn how to suggest structures to achieve one or more property objectives. In 2024, we published our first generative method of latent diffusion directly on top of the COATI embedding which uses classifier guidance (CG) to optimize one or more molecular properties (see the paper here: https://www.biorxiv.org/content/10.1101/2024.08.22.609169v1 or check out our GitHub (https://github.com/terraytherapeutics/COATI-LDM)). Recently, we have deployed a second-generation reinforcement learning-based generative method that uses a policy gradient (PG) algorithm. PG tends to generate more synthetically-accessible molecules than CG, with equivalent or better properties – and can directly leverage our predictive models for generation without needing separate diffusion guides per property.
Predict: EMMI’s prediction models assess ADME properties and potency across the proteome
With thousands to millions of molecules resulting from Generate tasks, it is very important to be able to profile these molecules to then select the few that will be synthesized. For this, we have a variety of curated Predict tasks that assess potency and selectivity against one or more targets, as well as non-potency properties such as ADME and physicochemical properties.
Since our experimentation is rooted in the largest database of binding data for small molecules, we opted to build global potency models from Day 1. We finished the first version of our core global potency prediction model, TerraBind, in 2023. Over the last 2 years, we’ve learned a lot and now deploy a unique two-tiered approach to potency prediction that allows us to evaluate thousands of times more molecules than the competition and thus more accurately pick the best ones for follow-up. Every single molecule generated is scored using an ultra-fast sequence-only model that predicts properties 20,000x cheaper and faster than public models, yielding a subset of promising molecules for further profiling. This subset is then scored for potency using a structure-based, multi-modal potency model that leverages a Pairformer representation derived from COATI3 and a protein LLM, but is still 20x cheaper and faster than public models. This speedup is due to our internal emphasis on “potency over pose” and results in both highly accurate potencies as well as useful poses that can be used for pocket ID or structure-based design.
The most promising molecules are further profiled by ADME and physicochemical models to predict the rest of the properties that need to be optimized for a molecule to move to clinical testing. These models include solubility, LogD, permeability, metabolism, clearance, and more. Collectively, EMMI’s generative and predictive models put thousands of suggested molecules up for consideration, all with predicted values across potency, selectivity, ADME, and physicochemical properties. This is, of course, too many molecules to make in any one DMTA cycle of drug discovery, which is why it is imperative to pair Generate and Predict with Select, to choose the best molecules for synthesis and testing.
Select: EMMI’s selection models utilize uncertainty quantification and more to select the optimal batch of molecules to synthesize
Indeed, in small molecule drug discovery, the question of which molecules to make and test in the next experimental cycle is undoubtedly one of the most consequential. Advanced program molecules typically require custom synthesis and each chemist can only make a few molecules per week. Picking the wrong set of molecules to synthesize can set a program back for many months or even years, increase costs significantly, and ultimately lead to the high failure rate that is endemic to drug discovery. Terray’s Generate and Predict models produce thousands of promising possibilities to pursue in every iteration, but the ultimate question where AI intersects with experimental reality is what is the optimal small set of compounds to synthesize and test?
It is tempting to be “greedy” and simply make the top few molecules based on a predicted property score – but this is a big mistake as the models have underlying uncertainty that is often correlated. For instance, models may highly value a particular structural motif that is present in all of the top compounds, and a greedy selection could exploit this artifact and result in molecules that are all failures. A common technique to reduce this risk is to consider both predicted score and structural diversity, but this is an indirect approach that often fails to incorporate model uncertainty that is shared between structurally different molecules.
Terray addresses this challenge with our newest model. EMMI’s Select model combines Epistemic Neural Networks with an acquisition function such as expected maximum (EMAX) to directly weight model uncertainty in synthesis selections. In retrospective benchmarking for potency optimization, this approach dramatically outperforms existing methods, with time and cost savings of 3x. For more information on our use of Epinets + EMAX to make optimal decisions for potency prediction and beyond, see our latest insight post and paper (https://arxiv.org/abs/2511.10590), or check out our GitHub (https://github.com/terraytherapeutics/terramax).
Working With EMMI Today and Tomorrow
After years of work building all of the components, we are excited to tell the world about EMMI! One of the keys to EMMI’s success is that scientists across Terray use it every day via an intuitive user interface to drive progress on all of our programs. This results in both constant platform improvement, and continual optimization of the integration between experimentation and machine intelligence. In one example of this, scientists are transitioning to working with EMMI through an agent. This means that instead of having to manually decide which models to employ and in which sequence, the scientists can now simply ask EMMI for the desired outcome and let EMMI plan the workflow itself. Most importantly, EMMI is delivering across a diverse pipeline with molecules that are completely novel in structure, addressing some of the most difficult challenges in drug discovery. We can’t wait to see EMMI’s innovations transform the lives of patients in need!